Numpy + Pandas
By: Matthew Qu & Asher Noel
Getting Started
Before we begin, we must first install the numpy and pandas libraries as they are not included in the standard Python library (check out our guide if you have any questions about installation). When we import these libraries, we typically abbreviate them as follows:
This isn't necessary, but since we'll be calling functions from these libraries so often, it saves us quite a bit of typing!
NumPy
Numpy (“Numerical Python”) is a general-purpose package that endows Python with efficient multi-dimensional array and matrix objects and operations.
Introduction
Python lists are important, but slow. Numpy array objects aim to be 50x faster than traditional Python lists. Unlike Python lists, Numpy array objects are stored at one continuous place in memory.
On its own, Numpy’s matrix operations are commonly used for linear algebra and as the biases of all scientific work. Often, other libraries take advantage of numpy’s speed by building atop it, such as PyTorch for machine learning and Pandas for data analysis.
Why is NumPy so fast?
Python’s dynamic typing makes vanilla Python slow. Every time Python uses a variable, it has to check that variable’s data type. Lists are arrays of pointers. Even when all of the referenced objects are of the same type, memory is dynamically allocated.
In contrast, Numpy arrays are densely packed arrays of a homogeneous numerical data type. This allows memory allocation to be continuous, and operations can parallelize.
Examples
Addition
Component-Wise Multiplicaiton
This is commonly referred to as a Hadamond product.
np.dot
With vectors, it is acceptable to use np.dot to calculate dot products. With matrices, numpy documentation says that it is preferable to use np.matmul():
Broadcasting
In math, dot product and matrix multiplication require the same dimensions. In numpy, this is not the case. Dimensions are compatible if they are 1) the same value or 2) one of them is 1. The size of the result array is the maximum along that particular dimension.
As a simple example, we will add a scalar to an array:
To read more about harder examples, check out the numpy documentation! For example, the dimension of adding a (3, 4, 5, 6) and (4, 6, 7) dimensional object will have dimension (3, 4, 5, 7).
Functions
Numpy comes with a trove of built-in functions, including np.matmul, np.zeros, np.arange, np.identity, and more. If you ever need to do something with a matrix, just check the documentation or google a query, and numpy can probably handle it!
Pandas
Introduction
Pandas is a powerful Python library that is specifically designed for data manipulation and analysis. Its name comes from the term panel data, which is data that contains information of individuals over a period of time.
Why use Pandas?
In general, Pandas makes it easy and intuitive to work with data; this includes cleaning, transforming, and analyzing data. Data from Pandas is also commonly used alongside other Python libraries such as SciPy, Matplotlib, and Scikit-learn for use in statistical analysis, data visualization, and machine learning, respectively.
NumPy and Pandas are almost always used in conjunction. In fact, Pandas is built on top of NumPy, and the two libraries work together internally as well. Because NumPy objects and operations are highly efficient, Pandas also executes very quickly.
Data Structures in Pandas
The two main data structures in pandas are Series and Dataframes. Series can be thought of as columns; they are
a one-dimensional array of values. We can create a Series by passing in an iterable to the argument data:
The first column is the index; by default, it is numerically indexed starting from 0. We can create custom indices by
setting the optional argument index equal to another iterable. This iterable must be of the same length as that
passed into the data argument. If data is a dictionary, the keys become indices and the values make up the data.
If Series are like columns, then Dataframes are like tables with both rows and columns. As with a Series, we can create a Dataframe from scratch:
For a DataFrame, the keys of the dictionary become column names, not indices. We'll be working with DataFrames most of
the time, but Series can arise when we extract data from a DataFrame. We can use the loc[] function to extract
data based on the label of the index:
Similarly, we can use the iloc[] function to extract data using numerical indices, not labels. This is useful for
when the indices are relabelled.
loc[] and iloc[] accept up to two arguments, the first being an expression that determines which rows to
extract, and the same being the same but for the columns (by default, all columns are selected). For example, we can use
slice notation to extract the entire
first column:
Importing and exporting data
We don't usually create DataFrames from scratch using dictionaries or lists - most of the time, we'll want to read external data stored in another file. Let's work with a real example. The data we'll be using comes from the U.S. Geological Survey of all earthquakes with magnitude 2.5 or greater that occurred on a randomly chosen day in 2020 (June 14). You can download the data here.
Pandas has a read_csv() function that automatically converts CSVs to DataFrames, using the first line as column
names:
tip
Pandas also has the functions read_json() and read_sql_query() to read data from JSON files and SQL
databases.
We can similarly export data from DataFrames to other files. This can be done using the to_csv() function (or
likewise the to_json() and to_sql() functions). For example, we can export the earthquakes DataFrame
(presumably after some changes) and save it under the name new_earthquakes:
Working with data in Pandas
We often work with data that contains an abundance of information. For large DataFrames, we can use the head()
function to examine the first five rows. We can also specify a different number of rows; for example, earthquakes.head(3)
would print only the first three rows. Similarly, the tail() function can be used for viewing the end of the DataFrame.
The output is still condensed, however, and we can see that there are actually 22 columns in our DataFrame. We can inspect the column names as follows:
tip
We can also use the info() function to get column names as well as some other useful data, such as how many
non-empty entries are in each column:
Let's index by id, but to avoid creating a new DataFrame, we set an argument inplace to be True. In
addition, let's extract just the first five columns because they contain the important data:
We can also sort by a specific column. Let's look at the most severe earthquakes for this day, so we want to sort by magnitude from highest to lowest:
We can also calculate the mean of rows and columns in the same way we would in numpy, specifying axis=0 to average
over all rows (leaving columns) and axis=1 to average over all columns (leaving rows). Specifically, let's
calculate the mean of the depth and mag columns, because taking the average of the other three wouldn't
give us much insight:
If we want to make changes to a row or column, we can use the apply() function. This function is used on a Series
and takes in a function to be applied on every element in the Series. For example, every entry in the time column
has a format similar to 2020-06-14T14:24:29.479Z. We can splice the string and get rid of the date and the extra
precision on the time:
Don't forget to update the DataFrame with the new time!
Pandas is also commonly used with other data visualization libraries. For example, we can use plotly to plot the locations of the epicenters on a world map:
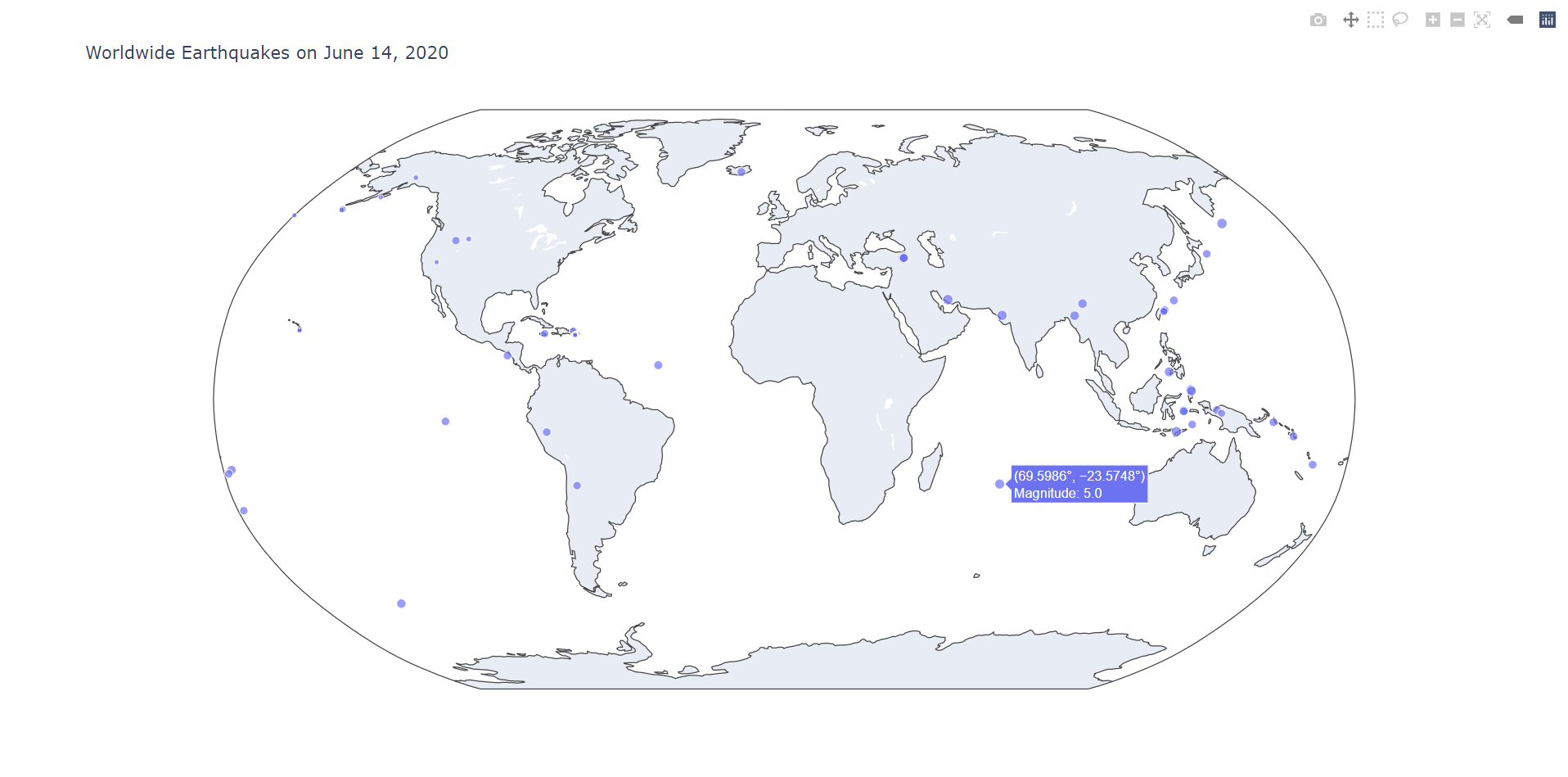
Hopefully this guide has served as an introduction to numpy and pandas as well as their widespread usefulness in data science.